
enclone can create these types of plots:
Honeycomb plots display cells as dots, with each clonotype represented as a hexagonal cluster of cells. enclone provides various controls over the configuration of the plots, as described in the next sections.
Hint.
You may wish to use the MIN_CELLS option
(see enclone help filter) to exclude tiny
clonotypes, which might otherwise crowd the image and slow down plotting.
enclone provides a number of ways to assign colors in such a plot. We describe them in order of precedence, i.e. color data for the first will be used if provided, etc.
The syntax for this is under development and fragmented at present.
PLOT="filename,origin1->color1,...,originn->colorn"Example:
enclone BCR=123085:123089 MIN_CELLS=10 PLOT="plot.svg,s1->blue,s2->red" NOPRINT
LEGEND=blue,123085,red,123089
Note the colon between 123085 and 123089. This tells enclone
that the two datasets are different origins from the same donor. For example, they might
represent cells collected at two time points. This is not actually true in this case, as
these two datasets have the same origin, but is needed to plot in this way.
The NOPRINT argument tells enclone to not generate its usual output, so
although the command itself will appear to do nothing, it will create a file
plot.svg.
If you're using a Mac, then the file plot.svg in the command can be displayed
by typing open plot.svg. In that case the application used to display the plot
will be picked for you. You can specify a particular app e.g. with
open -a "Google Chrome" plot.svg.

Here is a simpler example, which plots the clonotypes in a single dataset (plot not shown):
enclone BCR=123085 MIN_CELLS=10 PLOT="plot.svg,s1->blue" NOPRINT.
PLOT=filenameMETA
option (see enclone help input).
This assigns a color to each dataset.
PLOT specification, and assign a color to each barcode using
the BC option or the bc field for META.
PLOT_BY_ISOTYPE=filename. This plots by heavy chain
constant region name and
labels accordingly. (This only makes sense for BCR.) Some cells may be labeled "unassigned", for
one of three reasons: (1) no heavy chain was found; (2) no constant region was found; (3) two
heavy chains were found and had conflicting constant region names. Running with
MIN_CHAINS_EXACT=2
is usually a good idea to avoid noise coming from cells for which only a light chain was detected.
Currently a maximum of 12 constant region names is allowed. Let us know if you have more and we
will fix this. Note that PLOT_BY_ISOTYPE cannot be used with PLOT or
LEGEND.
Example: enclone BCR=123085,123089 MIN_CELLS=5 MIN_CHAINS_EXACT=2 NOPRINT
PLOT_BY_ISOTYPE=plot.svg

If desired, an additional argument
PLOT_BY_ISOTYPE_COLOR=color1,...,colorn
maybe used to define an alternate color list. The first color is for the undetermined case, and
the subsequent colors are in order by constant region name. enclone will fail if the list is
not long enough.
The legend may be suppressed using the argument SUPPRESS_ISOTYPE_LEGEND.
Example: enclone BCR=123085 MIN_CELLS=10 HONEY=out=plot.svg,color=var,u_cell1
NOPRINT
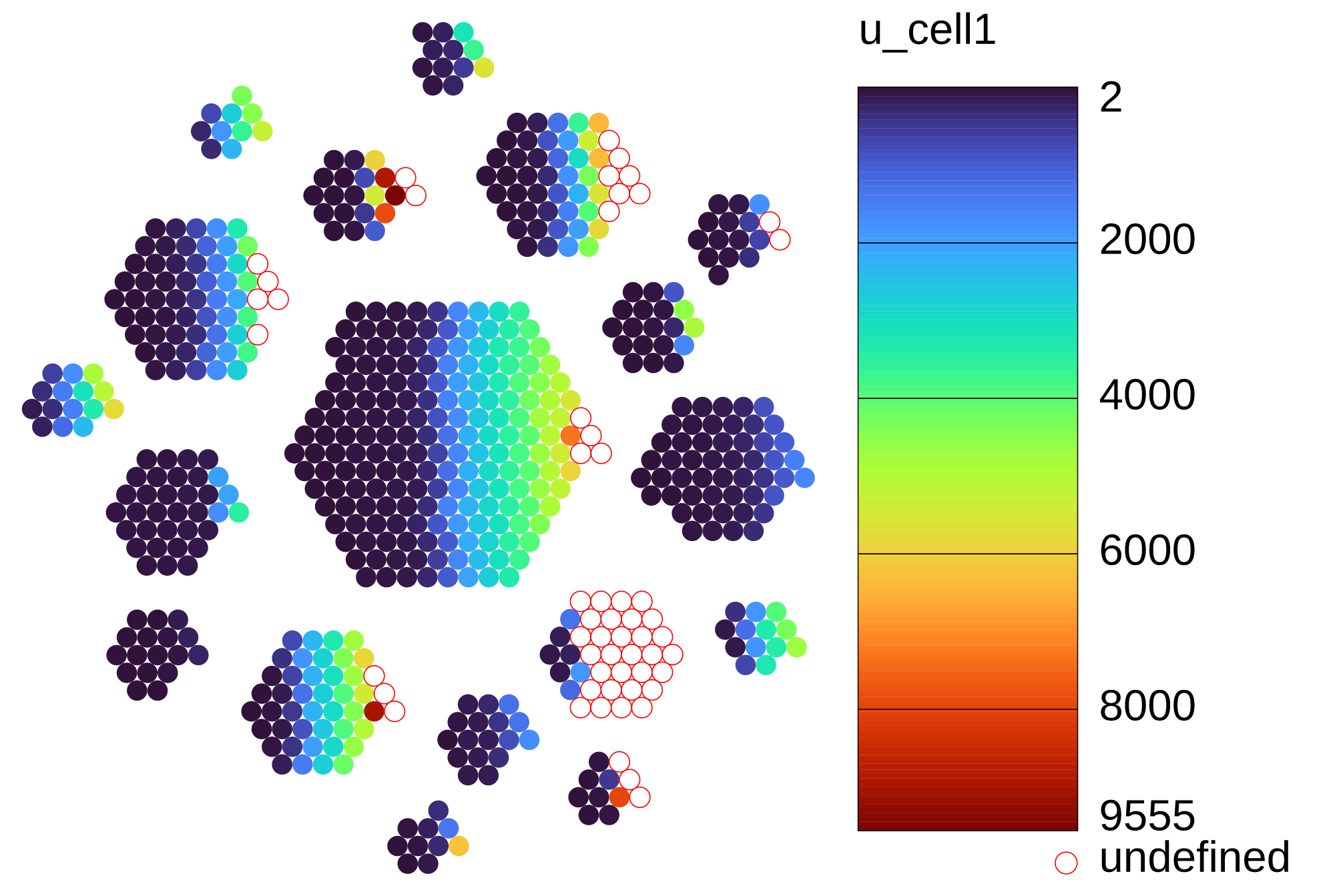
In the given dataset, for cells in clonotypes having at least ten cells,
the plot colors each by the value of the variable
u_cell1, the number of UMIs in the first chain within the cell's clonotype.
The variable is sometimes undefined because a clonotype can include cells which are missing the
first chain. Note also that using the variable u1 would have instead colored cells
by the median number of UMIs for their first chain, where the median is computed across the
exact subclonotype containing the cell.
See merged syntax, below.
Example: enclone BCR=123085,123089,124547 MIN_CELLS=5 HONEY=out=gui,color=dataset

See merged syntax, below.
10 categories are shown, reverse sorted by cell count. The last category is "other".
Example: enclone BCR=123085
HONEY=out=plot.svg,color=catvar,v_name1+v_name2,maxcat:10 NOPRINT CHAINS_EXACT=2

See merged syntax, below.
Here we show the syntax for coloring by variable or dataset.
Red fields are to be filled in by you.
| part | syntax | notes |
everything |
HONEY=out-spec,color-spec,legend-spec |
order of specification fields is ignored |
out-spec |
out=filename,width |
1. filename
is the output file to be generated; it should end with .svg
or .png depending on the desired output file type.
2. width is the width in pixels of the image
and may be omitted.
It only makes sense for .png files.The default value is 2000.
You could use, for example, a value of 4000 to get a higher resolution image.
|
legend-spec |
legend=none |
to suppress legend; omit this field to show the legend |
color-specdataset version |
color=dataset |
Specify coloring by dataset. The color scheme is fixed for now. |
color-specnumerical variable version |
color=var,abbr:name,turbo,scale-spec |
Specify coloring by numerical variable. 1. turbo is the name of the color map and may be omitted
2. abbr: may be omitted;
abbr is the display name
3. name is the name of the variable |
color-speccategorical variable version |
color=catvar,vars,maxcat:n |
Specify coloring by categorical variable. 1. vars
is the variable to be displayed; more than one variable may be used,
separated by +.
2. n is the maximum number of categories.
Categories are reverse ordered by number of cells and if there are too many categories, one
is lumped as "other" and displayed last.
|
scale-spec |
minmax,min,max
|
1. scale-spec may be omitted entirely2. min
or max or both may be omitted
3. They describe the range of values that define the color map. 4. Their default values are the min and max of the variable values. 5. If min or max is specified and a value is outside the range, the value
will be raised to min or lowered to max before assigning its color.
|
HONEY=out=plot.svg,color=var,u_cell1
HONEY=out=plot.svg,color=var,u_cell1,legend=none
HONEY=out=plot.svg,color=var,u1:u_cell1
HONEY=out=plot.svg,color=var,u_cell1,,minmax,0,10000
HONEY=out=plot.svg,color=dataset
The colors should be valid colors for use in an svg file. They can be named colors like red or
blue (see here for a full list)
or a hex specification like #00FFFF for aqua. The full color description for svg is
here.
enclone also recognizes the color abbreviations @1, ..., @6, which refer
to enclone's color blind friendly palette
(see enclone help color).
Each cell is shown as a disk having the given color, and each clonotype is shown as a cluster of these disks, which are positioned at random. The filename argument may be "stdout".
We describe here two options that can be used to modify the layout of honeycomb plots.
If desired, the honeycomb plots can be forced into the first quadrant using the
QUAD_HIVE option.
enclone BCR=123085:123089 PLOT="plot.svg,s1->blue,s2->red" QUAD_HIVE NOPRINT

enclone can make side-by-side honeycomb plots to facilitate comparison between different cell origins. For example, these origins could be from different tissues, or the cells could be prepared differently.
In this example we have two datasets, specified using a colon as 123085:123089,
which treats them as arising from two origins. These particular data are actually replicates.
Specification in this manner with SPLIT_PLOT_BY_ORIGIN causes all the
123085 cells to be shown in the left plot, and all the 123089 cells to
be shown in the right plot. Thus a cluster in the picture may be a partial clonotype, relative to
the entirety of the data.
In principle more than two origins can be specified.
enclone BCR=123085:123089 PLOT_BY_ISOTYPE=plot.svg SPLIT_PLOT_BY_ORIGIN NOPRINT

SPLIT_PLOT_BY_DATASET.
To add a legend to the graph, add the argument LEGEND to your command line. This
will give you an
auto-generated legend. You can also customize the legend by adding an argument of the form
LEGEND=color1,"text1",...,colorn,"textn"
to the command line.
When enclone creates a honeycomb plot, it tries to rearrange clonotypes so as to place
identically colored clonotypes next to each other. If you want to create two plots of the
same data, in which the positions of the cells are fixed by the first plot, you can do
this by providing an argument HONEY_OUT=filename to a first enclone command,
and then HONEY_IN=filename to a second enclone command, where both commands
refer to the same file.
If xvar and yvar are the names of variables
(see
enclone variable inventory,
enclone help lvars,
enclone help cvars and
enclone help parseable), and those variables
have numeric values, then
enclone can produce a plot of xvar versus yvar, showing a point
for each exact subclonotype (after all filtering) for which both variables are defined and are
numbers. The syntax is PLOTXY_EXACT="xvar,yvar,filename". The file is an
SVG file. We allow xvar = log10(v) for some parseable variable
v, and likewise for yvar. The quotes are only needed if the variables
have "funny" characters in them.
Example.
enclone BCR=123085 GEX=123217 NOPRINT PLOTXY_EXACT=HLA-A_g,CD74_g,plot.svg

An optional fourth argument sym may be added to force a square plot having
identical tic marks on the axes. This makes sense, for example, when comparing data from
replicates.
Given a list of numeric variables, enclone can display their pairwise correlation, where the correlation of two variables is the cosine similarity of their vectors (ranging across cells).
Example.
enclone BCR=123085 GEX=123217 SIM_MAT_PLOT=plot.svg,CDKN1A_g,CDKN1B_g,RBX1_g,IGLC1_g,IGLV3-21_g